From bench to bedside, a small step in cardiology?

Publishing your research, what is happening afterwards?
Suraya Elfrink and Annemiek van Spriel reflect on the past year, which was a beautiful and inspiring year also because of their publication in Blood, The journal of the American Society of Hematology. read the interviewPublishing your research, what is happening afterwards?
I was surprised by how many people actually read itPhD candidate Suraya Elfrink saw the research she conducted with Lotte de Winde and Michiel van den Brand published in Blood. Together with her supervisor Annemiek van Spriel, Suraya reflects on her research and the impact of being published.
Can you tell me a little about the research that was published in Blood, the journal of the American Society of Hematology?
Suraya: “Tetraspanin CD37 is frequently mutated in patients with diffuse large B cell lymphoma (DLBCL) occurring at immune-privileged sites. The CD37-mutated lymphoma B-cells show impaired CD37 cell surface localization, which has important implications for anti-CD37 immunotherapy. In other words, this study provides novel molecular insight into DLBCL at immune-privileged sites. It indicates that CD37-based therapies are more likely to be beneficial for DLBCL patients that harbor no CD37 mutations.”
Annemiek: “It’s, therefore, predictive for DLBCL patients who do have CD37 mutations as they don’t need to undergo the expensive and evasive anti-CD37 immunotherapy because we know that it will not help them. Practically, it means they can spend their time and energy on another treatment that will hopefully have a positive effect."
What is it about research that attracts you to do it Suraya: the fundamental questions or the possible impact on patients?
“I certainly like the fundamental questions,” says Suraya, "but I also see the personal picture concerning my research. I genuinely understand the impact of it. As a child, I always saw myself as an archeologist or perhaps a historian. Then I got cancer when I was around 13 years old. In eighth grade, you have to choose your subject package, and due to my illness, I decided to take subjects like Biology instead of History. I’ve never regretted that choice.
“I was treated right here at Radboudumc. One of the oncologists who treated me still works here. He knows I’m doing research as he saw an article about me on the back of the Radbode. He once asked how my PhD was going, and I’ll definitely send him my thesis when it’s done.”
Blood – which is a prestigious journal – published your PhD research. How did it come about, and what did getting published mean to you?
Suraya: “Annemiek and I submitted the article to Blood together. It was my first time submitting an article. I remember we sat behind the computer for 3 hours or something. I was yelling at the computer because technically things weren’t going the way they should. But Annemiek stayed remarkably calm.”
Annemiek: "Writing the article went smoothly. Blood was the first journal she submitted to, and after it was accepted, she revised the manuscript herself. I was so proud of her. She'd worked exceptionally hard. I enjoy working with PhD candidates: watching them work and develop into independent researchers. "
Suraya: "I understood that getting published in Blood was an accomplishment, but I didn't realize how big until the magazine came out, and people started congratulating me. I was surprised by how many people actually read it."
You do research independently, but how is the support within the research group?
Annemiek: "Our team consists of about ten people and includes postdocs, PhD candidates and Master's students. Everyone works hard and is motivated not just by their personal research but also each other's."
Suraya: “We’re a close team. There’s definitely no animosity. We all want to get published, and we help each other to get there. All successful publications are celebrated equally: there's no ranking. We have a 'Wall of Fame' within the Tumor Immunology department. When someone gets published, we post the article on that board. After ten posts, we have a drink together to celebrate as a department.
“Of course, getting published isn’t the most important thing. It’s about growing as researchers. We’re all passionate about our work. And our goal is to become independent researchers under Annemiek’s supervision. In the team, we can share our successes and our frustrations. We help remind each other that the failed experiments and annoying reviews will be worth it in the end.”
Annemiek: "I think the most important thing for research is to love what you do. Find a research topic that excites you. Then you don't have to stress."
Suraya, smiling: “Although you will be stressed!”
Annemiek, smiling too: “Yes, but you shouldn’t start off being stressed.”
What are your plans Suraya? Remain in research or do you have other ambitions?
Suraya: "I hope to get a postdoc position somewhere in the field of oncology, preferably in pediatric oncology. I want to go somewhere outside of the Netherlands. It would be useful to establish myself in a different environment and learn from that as well.
“I consciously chose not to work directly in the clinic. Considering my background – my ‘role as survivor’ – that would be too personal. Although, I do like reading stories about patients, via, for example, patient organizations. I enjoy the connection; it keeps me aware of what I'm doing it for.”

Cancer development and immune defense Nature Genetics
Rik Lindeboom et al. read moreCancer development and immune defense Nature Genetics
The impact of nonsense-mediated mRNA decay on genetic disease, gene editing and cancer immunotherapy
RIMLS researchers of the Faculty of Science developed a machine learning-based algorithm to predict which cancer patients benefit from immunotherapy.Mutations in our DNA can disrupt protein synthesis, sometimes causing truncated proteins which don't work as intended. Known as nonsense and frame shift mutations, these types of alterations can give rise to hereditary diseases and different types of cancer. To keep the number of truncated proteins to a minimum, human cells recognize and remove RNAs with nonsense mutations through a quality control process known as nonsense-mediated mRNA decay (NMD).
To better understand the effect of NMD on human disease, we built NMDetective, a tool describing every possible nonsense mutation that can occur in the human genome. Developed by large-scale statistical analyses based on machine learning, the algorithm identifies which mutations in the genome are susceptible to NMD. Using NMDetective to analyze thousands of genetic variants that give rise to hereditary diseases in humans, revealed that NMD activity often leads to a greater severity of the disease. This suggests that pharmacological NMD inhibition could slow the progression of many different genetic diseases. We also discovered that NMD activity is important for the prediction of successful outcome of immunotherapy in cancer. While cancer cells contain many mutations which can be recognized by the immune system to destroy them, we show that NMD often suppresses their presentation to immune cells. Our algorithm can therefore help identifying the patients that will benefit from these therapies, providing a potential tool for personalized medicine in cancer.

Legend
Graphical illustration of predictive decision trees that are optimized to decide if a cancer mutation will become visible to the immune system. For every branch in each tree, the machine learning algorithm asks a question about the genomic environment of a cancer mutation, starting at the bottom and eventually placing each mutation in a leaf that is orange or blue. Mutations that end up in blue leafs will be destroyed by the quality control process NMD, but mutations in orange leaves will be left untouched and can therefore be seen by the immune system of the patient, making them more responsive to cancer immunotherapy.
Lindeboom RGH, Vermeulen M, Lehner B, Supek F. Nat Genet. 51, 1645-1651, 2019

Infectious diseases and global health Science Translational Medicine
Martin Jaeger et al. read moreInfectious diseases and global health Science Translational Medicine
SIGLEC15 is a novel receptor in the immune response against C. albicans and genetic alterations contribute to the pathogenesis of recurrent vaginal infections
Most women will experience a least one vaginal infection in their life time and a small proportion of around 8% will experience recurrent episodes, called RVVC (recurrent vulvovaginal candidiasis). Some predisposing factors are known, such as diabetes, just as long‑term use of antibiotics or contraceptive pills, but it is unclear why some women which are not affected by those risk factors are more sensitive to RVVC than others. We hypothesized that a genetic component is the pathogenesis of RVVC was most likely. By genetically screening two independent cohorts of patients and controls and combining these data with immunological datasets, we identified SIGLEC15 as a novel receptor involved in both Candida, recognition as well as in the pathogenesis of RVVC. Our human data suggest that a specific genetic alteration in the SIGLEC15 gene leads to increased levels of cytokines rather than to a shortage of cytokines. This “hyper-inflammation” contributes to the symptoms seen in RVVC patients. To confirm this, we used a mouse vaginal infection model and obtained similar results. We show that Candida is able to induce SIGLEC15 expression and that specific silencing of SIGLEC15 at the vaginal surface led to an increase in Candida colonization accompanied with altered cytokine levels. Finally, these results not only increase our understanding of the RVVC pathogenesis, but might also lead to the development of directed therapies in women suffering from RVVC.
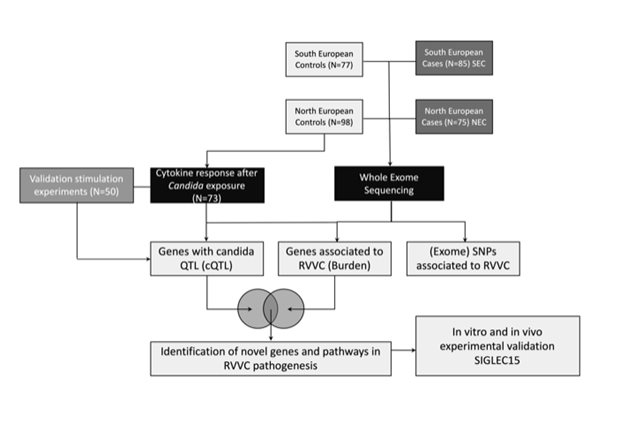
Legend
Schematic overview of the study. Two independent cohorts of RVVC patients and controls underwent exome-sequencing. For a subset of healthy controls Candida-induced cytokine data were generated. After overlapping significant hits of both genetic and immunological analyses, we identified novel pathways and genes being dysregulated in RVVC. One of these genes was SIGLEC15, which was experimentally validated in vivo and in vitro.
Jaeger M, Pinelli M, Borghi M, Constantini C, Dindo M, van Emst L, Puccetti M, Pariano M, Ricaño-Ponce I, Büll C, Gresnigt MS, Wang X, Gutierrez Achury J, Jacobs CWM, Xu N, Oosting M, Arts P, Joosten LAB, van de Veerdonk FL, Veltman JA, Ten Oever J, Kullberg BJ, Feng M, Adema GJ, Wijmenga C, Kumar V, Sobel J, Gilissen C, Romani L, Netea MG. Sci Transl Med. 11, eaar3558, 2019.

Inflammatory diseases Science Translational Medicine
Joost Schalkwijk et al. read moreInflammatory diseases Science Translational Medicine
Antimalarial pantothenamide metabolites target acetyl–coenzyme A biosynthesis in Plasmodium falciparum
A decade ago, the Experimental Dermatology lab of the Radboudumc started to develop anti-psoriasis drugs, based on the identification of a target epidermal enzyme involved in coenzyme A metabolism. Although a number of potent enzyme inhibitors were developed, the psoriasis drug discovery program came to a dead end. Several other applications of these compounds were then considered, including metabolic and infectious diseases. In a collaborative effort with the department of Medical Microbiology and the Radboudumc spin-off TropIQ Health Sciences, the development of promising lead compounds, called pantothenamides, was continued for malaria, one of the most deadly infectious diseases of our time. As there is no effective vaccine for malaria, and resistance against current treatments is on the rise, the development of new antimalarial drugs is a high priority. A Nijmegen-led international consortium of malaria researchers generated and characterized novel pantothenamides that showed favorable characteristics for clinical development. A humanized mouse model for malaria suggests that the administration of a single dose of a candidate drug may cure the disease completely. Additionally, it also stops the transmission of the malaria parasite from humans to mosquitoes, and the chemistry of the compounds is straightforward and cheap. The molecule has a mechanism of action that is distinct from currently marketed antimalarials which implies that that there is no resistance to the drug yet. A clinical candidate has been selected and is now taken through the next stages of development to determine safety, followed by first-in-human clinical studies. The research that led to this candidate drug has been published in Science Translational Medicine.
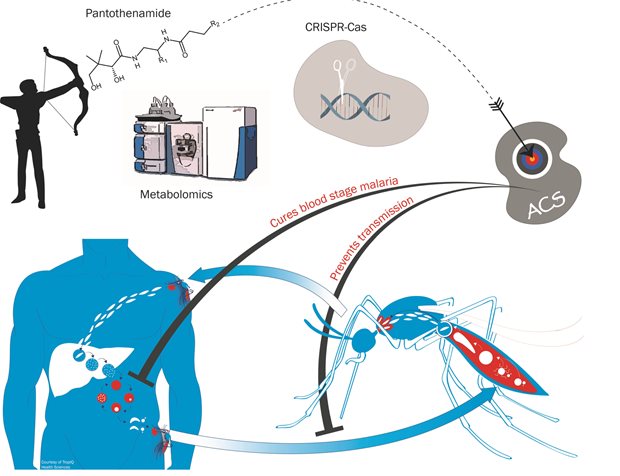
Legend
We generated stable pantothenamides with potent, low nanomolar activity against malaria parasites. Using a combination of metabolomics and gene editing technology in Plasmodium falciparum, we elucidated the mechanism of action of these compounds. By virtue of their similarity to endogenous pantothenate metabolites, pantothenamides are converted to antimetabolites by the cellular coenzyme A biosynthetic machinery. These antimetabolites interfere with enzymes involved in acyl-coenzyme A biosynthesis such as acetyl-coenzyme A synthetase (ACS). Pantothenamides are active against asexual blood stages of the parasite but also against gametocytes, thereby blocking transmission of the disease from the host to the mosquito.
Schalkwijk J, Allman EL Jansen PAM, de Vries LE, Verhoef JMJ, Jackowski S, Botman PNM, Beuckens-Schortinghuis CA, Koolen KMJ, Bolscher JM, Vos MW, Miller K, Reeves SA, Pett H, Trevitt G, Wittlin S, Scheurer C, Sax S, Fischli C, Angulo-Barturen I, Jiménez-Diaz MB, Josling G, Kooij TWA, Bonnert R, Campo B, Blaauw RH, Rutjes FPJT, Sauerwein RW, Llinás M, Hermkens PHH, Dechering KJ. Sci Transl Med. 11, eaas9917, 2019.

Mitochondrial diseases Nucleic Acids Research
Fenna Hensen et al. read moreMitochondrial diseases Nucleic Acids Research
Mitochondrial RNA granules are critically dependent on mtDNA replication factors Twinkle and mtSSB
Mitochondrial DNA (mtDNA) requires a unique and specialized gene expression system for its replication, transcription and translation, involving 200-300 nuclear-encoded gene-products. Therefore, mitochondrial function depends on the integration of few critical mtDNA-encoded factors together with many nuclear factors. Newly synthesized mitochondrial RNA has been observed to accumulate in discrete structures within the mitochondrial network, so-called RNA granules. These structures have been proposed to function as RNA processing and ribosome assembly factories, but were also shown to function in RNA degradation. In the current paper we could show that two classical mtDNA replication factors, the mtDNA helicase, Twinkle and the single-stranded DNA-binding protein, mtSSB have an unanticipated role in RNA granule biology. Transient Twinkle depletion led to a rapid disappearance of RNA granules but not overall mitochondrial RNA synthesis. Twinkle was furthermore shown to partially co-localize with RNA granules (see Figure). Transient mtSSB depletion on the other hand was shown to stabilize RNA granules and result in an mtRNA degradation defect. These results again showed moonlighting functions for well characterized mitochondrial proteins but also suggested that the principal role of RNA granules lies in degrading non-coding RNA species encoded by mitochondrial DNA. As both Twinkle and mtSSB are mitochondrial disease genes, the results also show that disease phenotypes should be considered beyond their role in mtDNA replication.
Legend
Transient Twinkle depletion results in rapid loss of RNA granules. U2OS cells were transiently depleted of Twinkle. Shown here are 20x30 µm representative subcellular sections of Control or Twinkle siRNA treated U2OS cells and stained for Twinkle, GRSF1 and mtDNA. GRSF1 is one of the classical markers for RNA granules, in control cells showing intense puntate structures (RNA granules) and a weak uniform mitochondrial fluorescence. Twinkle co-localizes with these structures that are in addition found in close association with mtDNA. Twinkle depletion is shown here to deplete GRSF1 punctae. Likewise GRSF1 depletion, results in loss of Twinkle punctae (see manuscript), suggesting an intimate relationship between Twinkle and GRSF1 in RNA granule biology.
Hensen F, Potter A, van Esveld SL, Tarrés-Solé A, Chakraborty A, Solà M, Spelbrink JN. Nucleic Acids Res. 47, 3680-3698, 2019.

Nanomedicine Nature Communications
Koen van den Dries et al. read moreNanomedicine Nature Communications
A structural framework for micron-sized cellular drills
Cells of the immune system such as antigen-presenting dendritic cells (DCs) migrate long distances through the body, thereby crossing many tissue boundaries. Although it was known that this specialized migration process is controlled by micron-sized cellular structures called podosomes, an adequate structural framework for podosomes was still missing. Here, we used a variety of super-resolution microscopy techniques to study the nanoscale architecture of podosomes in human DCs. We found that individual podosomes comprise functionally distinct, nanoscale substructures defined by different actin organizations. A central branched actin core is encased by a linear actin dome to generate mechanical forces for cell protrusion. The dome is linked to the plasma membrane by rope-like actin filaments to sense differences in tissue stiffness. Lastly, neighboring podosomes are connected by actin filaments to redisribute cellular forces. We established that, when exposed to a stiff environment, podosomes mediate long-range substrate exploration, associated with degradative behavior, whereas on soft material, podosomes display only short-range connectivity and a protrusive, non-degradative state. Our studies provide a comprehensive structure-function relationship for nanoscale podosome architecture and greatly furthers our understanding of how immune cells detect weak spots in basement membranes to cross tissue boundaries. Importantly, since podosomes are related to protrusive cancer cell invadopodia, our work may also provide novel leads to improve anti-cancer therapies.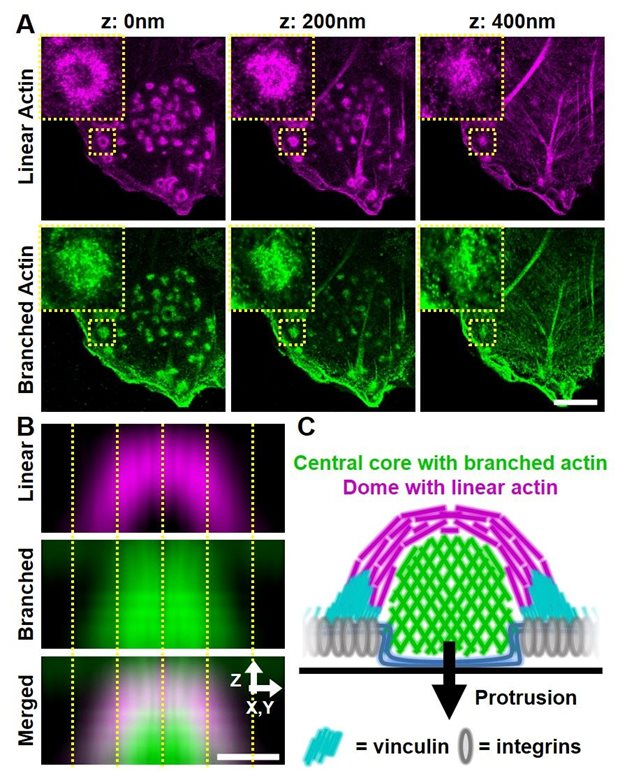
Legend
A) 3D super-resolution image of DCs labelled for linear (magenta) and branched (green) actin (Zeiss Airyscan, microscopy imaging center, Radboudumc). The insets show individual podosomes. B) Averaged side view of many podosomes, showing the linear dome and the branched core within single podosomes. The scale bar is 500 nm. C) Schematic depiction of the substructures within a single podosome, also including the adaptor protein vinculin and the protruded plasma membrane (blue).
van den Dries K, Nahidiazar L, Slotman JA, Meddens MBM, Pandzic E, Joosten B, Ansems M, Schouwstra J, Meijer A, Steen R, Wijers M, Fransen J, Houtsmuller AB, Wiseman PW, Jalink K, Cambi A. Nat Commun. 10, 5171, 2019.

Rare cancers Scientific Reports
Sandra Heskamp et al. read moreRare cancers Scientific Reports
Non-invasive imaging of hypoxic tumor areas in head and neck cancer
Hypoxia is a common feature of solid tumors and is associated with poor outcome in cancer patients. Tumor areas located distant from blood vessels are exposed to low oxygen levels which results in substantial cellular stress causing tumor cells to either die or adapt to these harsh conditions. Hypoxia-adapted tumor cells acquire a phenotype that is highly aggressive, metastatic, and therapy resistant. Therefore, patients with hypoxic tumors are less likely to respond to different types of therapy, including radiotherapy, chemotherapy, and immunotherapy. In order to improve patient outcome, it is essential to accurately characterize hypoxic tumor areas and if necessarily, adjust treatment accordingly. One key feature of chronic tumor hypoxia is the upregulation of carbonic anhydrase IX (CAIX). In this publication, we aimed to develop a novel non-invasive imaging technique to detect hypoxic tumor subvolumes in head and neck cancer xenografts. We have compared different radiolabeled CAIX-targeting molecules (antibody, antibody F(ab)’2 fragment, and affibody) for their potential to image CAIX-positive tumor areas by SPECT imaging. All three radiotracers were able to differentiate between tumors with high versus low CAIX expression. However, the antibody showed highest absolute tumor uptake. Future studies will be performed to assess whether it is feasible to develop this imaging further to use in patients with head and neck cancer.
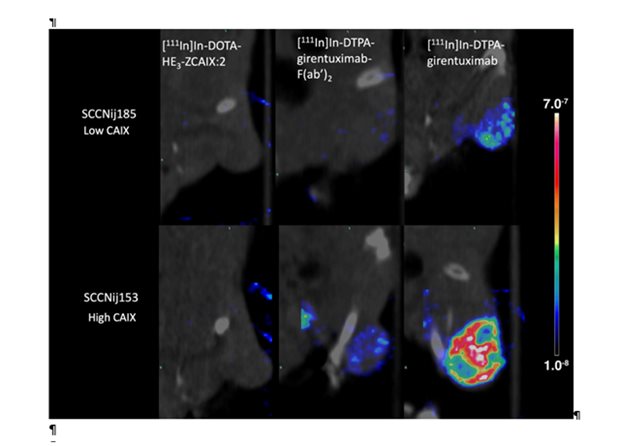
Legend
SPECT/CT scans of mice with head and neck cancer tumors, injected with radiolabeled affibody ([111In]In-DOTA-HE3-ZCAIX:2), antibody F(ab’)2 fragments ([111In]In-DTPA-girentuximab-F(ab’)2), or intact antibody ([111In]In-DTPA-girentuximab). These scans demonstrate the capacity of the different radiotracers to discriminate between tumor with high versus low CAIX-expressing tumor areas.
Huizing FJ, Garousi J, Lok J, Franssen G, Hoeben BAW, Frejd FY, Boerman OC, Bussink J, Tolmachev V, Heskamp S. Sci Rep. 9, 18898, 2019

Reconstructive and regenerative medicine Proc Natl Acad Sci U S A
Jeroen van den Beucken et al. read moreReconstructive and regenerative medicine Proc Natl Acad Sci U S A
2-stage reconstruction of the lower jaw
Tissue defects require surgical if spontaneous healing is not possible. For bone defects, the size largely determines the treatment options, of which autologous bone transplantation is preferred. However, bone defects in complex anatomical bones, such as the lower jaw, require an approach that warrants autologous vascularized bony tissue of customized geometry.
Here, reconstruction of a lower jaw bone defect was successfully achieved by a 2-stage approach using a sheep model. In stage 1, a mandibular bone defect was created simultaneously with the placement of a mold containing autologous bone (= bioreactor) with equal dimensions against a rib. Further, a polymeric device was implanted in the lower jaw bone defect to promote healing of the soft tissue envelop. After 9 weeks, stage 2 was performed, which included removal of the polymeric device, isolation of the bony transplant with blood vessel connection from the bioreactor, and transplantation of it to the lower jaw bone defect site. After an additional 12 weeks of healing, this 2-stage reconstruction demonstrated to be successful in 83% of reconstructed lower jaws. Given that clinically available biomaterials were used, this strategy justifies rapid clinical translation to improve outcomes in patients with large lower jaw bone defects.
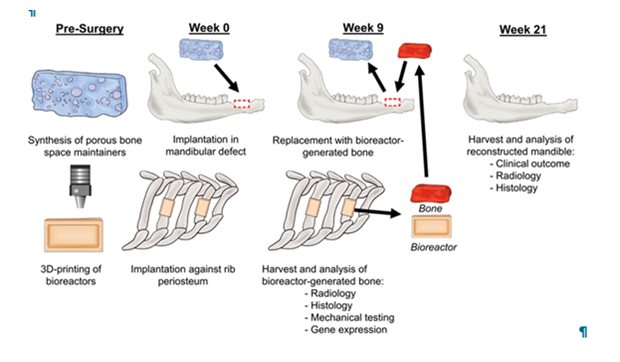
Legend
Schematic overview of experimental approach. Polymeric space maintainers and bioreactors were prepared in advance. At week 0, a bone defect was created in the lower jaw, into which a space maintainer was placed to enhance healing of a soft tissue envelope; bioreactors filled with autologous bone were implanted against the rib(s) of the same animal. At week 9, the vascularized bone from the bioreactors was retrieved, the space maintainer removed, and the vascularized bone graft was implanted into the jaw bone defect and anastomosed to the blood circulation. At week 21, mandibles were harvested to evaluate clinical outcome and analyse bone formation via radiological and histological techniques.
Tatara AM, Koons GL, Watson E, Piepergerdes TC, Shah SR, Smith BT, Shum J, Melville JC, Hanna IA, Demian N, Ho T, Ratcliffe A, van den Beucken JJJP, Jansen JA, Wong ME, Mikos AG. Proc Natl Acad Sci U S A. 116, 6954-6963, 2019.

Renal disorders Gastroenterology
René van Aerts et al. read moreRenal disorders Gastroenterology
Lanreotide Makes Its Mark in Tx of Rare Liver Disease
Polycystic liver disease (PLD) with large livers is the most common extra-renal manifestation in autosomal dominant polycystic kidney disease (ADPKD). Short term studies have shown that Lanreotide, which is a somatostatin analogue, reduces liver volume in these patients. The evidence for prolonged efficacy of Lanreotide was lacking. We performed an open-label, randomized trial in 175 ADPKD patients suffering from ADPKD using monthly Lanreotide (n=93) administered or standard of care (n=82) for 120 weeks. We found that Lanreotide reduced the combined, height-adjusted liver and kidney volume by 7.18%, an effect that was still present four months after the last injection of Lanreotide. An earlier study by the same researchers in the same population had shown that Lanreotide treatment did not affect the decline of renal function in this group. The conclusion is that for ADPKD patients with symptomatic PLD, long-term treatment with Lanreotide should be considered to halt the growth of polycystic liver and kidneys.
Legend
Mild, moderate and severe phenotype according to the Kim classification. In all three images the liver is accentuated; cysts are colored dark, whereas the lighter parts resemble normal liver parenchyma. The panels indicate coronal CT image (with contrast) with three-dimensional view of mild (A), moderate (B) and severe (C) phenotype.
van Aerts RMM, Kievit W, D'Agnolo HMA, Blijdorp CJ, Casteleijn NF, Dekker SEI, de Fijter JW, van Gastel M, Gevers TJ, van de Laarschot LFM, Lantinga MA, Losekoot M, Meijer E, Messchendorp AL, Neijenhuis MK, Pena MJ, Peters DJM, Salih M, Soonawala D, Spithoven EM, Visser FW, Wetzels JF, Zietse R, Gansevoort RT, Drenth JPH; DIPAK-1 Investigators. Gastroenterology. 157, 481-491, 2019.

Urological cancers Oncogene
Vicky Luna Velez et al. read moreUrological cancers Oncogene
Suppression of prostate tumor cell survival by antisense oligonucleotide-mediated inhibition of AR-V7 mRNA synthesis
One of the mechanisms by which advanced prostate cancer develops resistance to androgen deprivation therapy is the elevated expression of C-terminally truncated androgen receptor (AR) variants. These variants, such as AR-V7, originate from aberrant splicing of the AR pre-mRNA and the inclusion of a cryptic exon containing a premature stop codon in the mRNA. The resulting loss of the ligand-binding domain allows AR-V7 to act as a constitutively active transcription factor. Here, we designed two antisense oligonucleotides (AONs) directed against cryptic splicing signals within the AR pre-mRNA. These two AONs, AON-ISE and AON-ESE, demonstrated high efficiency in silencing AR-V7 splicing without affecting full-length AR expression. The subsequent downregulation of AR-V7-target gene UBE2C was accompanied by inhibition of androgen-independent cell proliferation and induction of apoptosis in castration-resistant prostate cancer (CRPC)-derived cell line models 22Rv1, DuCaP, and VCaP. Our results show that splicing-directed AONs can efficiently prevent expression of AR-V7, providing an attractive new therapeutic option for the treatment of CRPC.
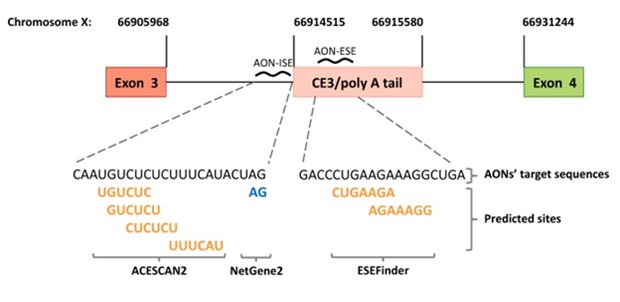
Legend
Design of antisense oligonucleotides (AONs). Schematic representation (not to scale) of the AONs designed to prevent splicing of androgen receptor (AR) pre-mRNA into AR-V7 mRNA. AON-ISE is complementary to the intronic splicing enhancer (ISE) sites predicted by ACESCAN2, and the cryptic “GA” splice acceptor dinucleotide motif, predicted by NetGene2. AON-ESE is complementary to the region harboring the ESEfinder-predicted exonic splicing enhancer (ESE) sites. Predicted splicing enhancer sites are bold and yellow, and the predicted cryptic splice acceptor site is on blue. The corresponding genomic coordinates (Human Genome Assembly February 2019, HG19) are marked by vertical lines pointing at the 5′ and/or 3′ junctions of exon 3, cryptic exon 3 (CE3), and exon 4
Luna Velez MV, Verhaegh GW, Smit F, Sedelaar JPM, Schalken JA. Oncogene. 38, 3696-3709, 2019.

Vascular damage New England Journal of Medicine
Niels van Royen et al. read moreVascular damage New England Journal of Medicine
Coronary angiography after cardiac arrest without ST-segment elevation
Ischemic heart disease is a major cause of out-of-hospital cardiac arrest. The role of immediate coronary angiography and percutaneous coronary intervention (PCI) in the treatment of patients who have been successfully resuscitated after cardiac arrest in the absence of ST-segment elevation myocardial infarction (STEMI) remains uncertain.
Niels van Royen and colleagues, theme Vascular damage, found out that among patients who had been successfully resuscitated after out-of-hospital cardiac arrest and had no signs of STEMI, a strategy of immediate angiography was not better than a strategy of delayed angiography with respect to overall survival at 90 days. This has major implications for the logistics and initial strategy in these patients. Also, it underlines that focus of future research should be on issues like early brain protection, cooling and response times.
In a multicenter trial, they randomly assigned 552 patients who had cardiac arrest without signs of STEMI to undergo immediate coronary angiography or coronary angiography that was delayed until after neurologic recovery. All patients underwent PCI if indicated. The primary end point was survival at 90 days. Secondary end points included survival at 90 days with good cerebral performance or mild or moderate disability, myocardial injury, duration of catecholamine support, markers of shock, recurrence of ventricular tachycardia, duration of mechanical ventilation, major bleeding, occurrence of acute kidney injury, need for renal-replacement therapy, time to target temperature, and neurologic status at discharge from the intensive care unit.
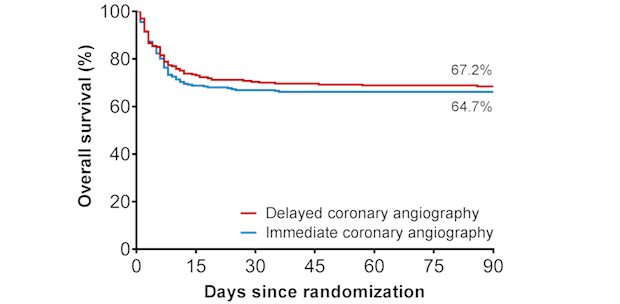
Legend
In patients with out-of-hospital-cardiac-arrest survival is not improved if an immediate angiography and additional coronary intervention is performed.
Lemkes JS, Janssens GN, van der Hoeven NW, Jewbali LSD, Dubois EA, Meuwissen M, Rijpstra TA, Bosker HA, Blans MJ, Bleeker GB, Baak R, Vlachojannis GJ, Eikemans BJW, van der Harst P, van der Horst ICC, Voskuil M, van der Heijden JJ, Beishuizen A, Stoel M, Camaro C, van der Hoeven H, Henriques JP, Vlaar APJ, Vink MA, van den Bogaard B, Heestermans TACM, de Ruijter W, Delnoij TSR, Crijns HJGM, Jessurun GAJ, Oemrawsingh PV, Gosselink MTM, Plomp K, Magro M, Elbers PWG, van de Ven PM, Oudemans-van Straaten HM, van Royen N. N Engl J Med. 380, 1397-1407, 2019.

Womens cancers Scientific Reports
Altuna Halilovic et al. read moreWomens cancers Scientific Reports
HER2, chromosome 17 polysomy and DNA ploidy status in breast cancer: a translational study
Breast cancer treatment depends on human epidermal growth factor receptor-2 (HER2) status, which is often determined using dual probe fluorescence in situ hybridisation (FISH). Hereby, also loss and gain of the centromere of chromosome 17 (CEP17) can be observed (HER2 is located on chromosome 17). CEP17 gain can lead to difficulty in interpretation of HER2 status, since this might represent true polysomy. With this study we investigated whether polysomy is present and how this effects HER2 status in six breast cancer cell lines and 97 breast cancer cases, using both immunohistochemical and molecular tests. We observed no isolated polysomy of chromosome 17 in any cell line. However, FISH analysis did show CEP17 gain in five of six cell lines, which reflected gains of the whole chromosome in metaphase spreads and aneuploidy with gain of multiple chromosomes in all these cases. This was also seen in patients' samples, in which gain of CEP17 correlated with aneuploidy of the tumor (91.1%; p<0.001). Our results indicate that CEP17 gain is not due to isolated polysomy, but rather due to widespread DNA aneuploidy. As aneuploidy is associated with poor clinical outcome, irrespective of tumor grade, this could improve future therapeutic decision making.
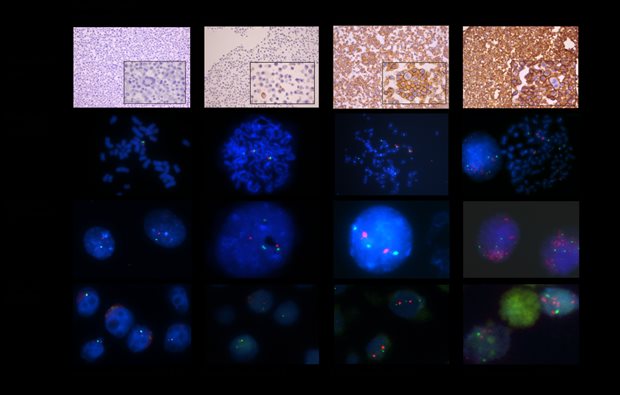
Legend
Examples of HER2 IHC of agarcyto slides and HER2/CEP17 FISH results on metaphase spreads, interphase nuclei, and agarcyto slides. Due to focusing (*) or cutting artefacts (**), not all of the spots of CEP17 (green) or the HER2 gene (red) may be visible.***: Partial loss of the HER2 gene. ****: HER2 gene amplified.
Halilovic A, Verweij DI, Simons A, Stevens-Kroef MJPL, Vermeulen S, Elsink J, Tops BBJ, Otte-Höller I, van der Laak JAWM, van de Water C, Boelens OBA, Schlooz-Vries MS, Dijkstra JR, Nagtegaal ID, Tol J, van Cleef PHJ, Span PN, Bult P. Sci Rep. 9, 11679, 2019.