20 August 2020
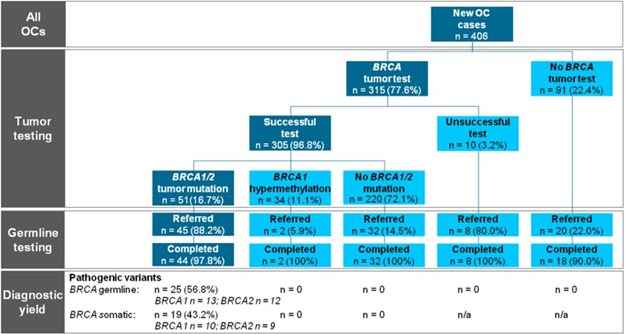
Figure 1.
Flowchart of the study showing the preferred universal tumor BRCA1/2 workflow on the left side in the dark boxes. n = 0 indicates that the test has been performed and no mutation was present. n = n/a indicates the test has not been performed or the test was unsuccessful and no result could be obtained. OC = epithelial ovarian cancer.
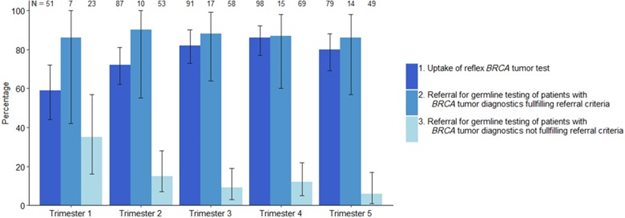
Figure 2.
Uptake of universal BRCA1/2 tumor testing and referral for genetic predisposition testing over time. Error bars represent 95% confidence intervals.

Background
Women with epithelial ovarian cancer (OC) have a higher chance to benefit from poly (ADP-ribose) polymerase inhibitor (PARPi) therapy if their tumor has a somatic or hereditary BRCA1/2 pathogenic variant. Current guidelines advise BRCA1/2 genetic predisposition testing for all OC patients, though this does not detect somatic variants. We assessed the feasibility of a workflow for universal tumor DNA BRCA1/2 testing of all newly diagnosed OC patients as a prescreen for PARPi treatment and cancer predisposition testing.Methods
Formalin-fixed paraffin-embedded tissue was obtained from OC patients in seven hospitals immediately after diagnosis or primary surgery. DNA was extracted, and universal tumor BRCA1/2 testing was then performed in a single site. Diagnostic yield, uptake, referral rates for genetic predisposition testing, and experiences of patients and gynecologists were evaluated.Results
Tumor BRCA1/2 testing was performed for 315 (77.6%) of the 406 eligible OC samples, of which 305 (96.8%) were successful. In 51 of these patients, pathogenic variants were detected (16.7%). Most patients (88.2%) went on to have a genetic predisposition test. BRCA1/2 pathogenic variants were shown to be hereditary in 56.8% and somatic in 43.2% of patients. Participating gynecologists and patients were overwhelmingly positive about the workflow.Conclusions
Universal tumor BRCA1/2 testing in all newly diagnosed OC patients is feasible, effective, and appreciated by patients and gynecologists. Because many variants cannot be detected in DNA from blood, testing tumor DNA as the first step can double the identification rate of patients who stand to benefit most from PARP inhibitors.
Figure 1.
Flowchart of the study showing the preferred universal tumor BRCA1/2 workflow on the left side in the dark boxes. n = 0 indicates that the test has been performed and no mutation was present. n = n/a indicates the test has not been performed or the test was unsuccessful and no result could be obtained. OC = epithelial ovarian cancer.

Figure 2.
Uptake of universal BRCA1/2 tumor testing and referral for genetic predisposition testing over time. Error bars represent 95% confidence intervals.
Related news items

Pathology reporting of gastric endoscopic resections: Recommendations from the International Collaboration on Cancer Reporting
17 November 2021 Iris Nagtegaal, theme Tumours of the digestive tract, and colleagues published her research project in Gastroenterology about pathological reporting of gastric endoscopic resections. go to page
Chella van der Post and Francesco Ciompi received a grant from Hanarth Foundation
11 November 2021 Unmasking the invisible cancer: digital detection of diffuse-type gastric carcinomas. go to page
RIMLS awards call for nominations
19 October 2021 RIMLS awards several prizes to stimulate and honor our (young) researchers. Upcoming awards are Supervisor of the Year, Best Master Thesis, Best Publication, Best Image and more. Send your nominations now before 24 November 2021. go to page
Young people with colorectal cancer deserve their own treatment
27 July 2021 Richarda de Voer and Marjolijn Ligtenberg's study on clinical, pathological, genetic and molecular characteristics of colorectal tumors in adolescents and adults 25 years of age or younger is published in Clinical Gastroenterology and Hepatology. go to page
The international dataset for colorectal local excisions is published
16 June 2021 Iris Nagtegaal and colleagues presented their first paper on the colorectal cancer dataset and published this in the journal Gastroenterology. go to page
